Abstract
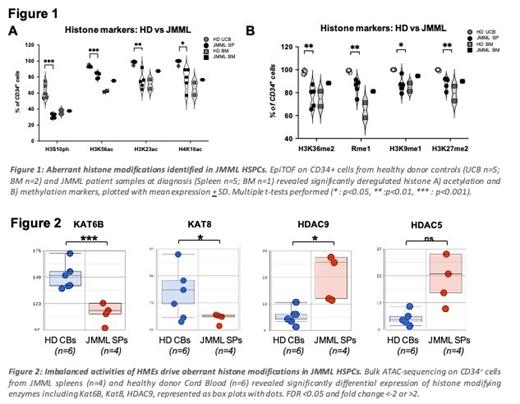
Juvenile myelomonocytic leukemia (JMML) is a deadly pediatric cancer characterized by excessive accumulation of monocytes/macrophages with myelodysplastic features and splenomegaly. Hematopoietic stem cell transplantation (HSCT) is the only curative option, but relapse occurs in ~50% of the patients. Mutations in the RAS pathway are the major drivers and DNA hypermethylation is associated with severe clinical phenotype. DNA methylation is linked to histone modifications in normal cell development and cancer pathogenesis. Gene expression levels are programmed by histone modifications. However, a comprehensive characterization of histone modifications in JMML is yet to be elucidated. It was reported that the CD34+ hematopoietic stem and progenitors cells (HSPCs) possess leukemia initiation potential (Caye et al., 2015; Krombholz et al., 2016; Louka et al., 2021; Yoshimi et al., 2017). We hypothesized that dysregulated histone modifications contribute to leukemogenesis in JMML. To this goal, we investigated epigenetic landscape at the single-cell level in JMML HSPCs using cytometry by time-of-flight (EpiTOF). We isolated HSPCs from the spleens of JMML patients with PTPN11 mutations which were collected at diagnosis, before treatment (provided by the European Working Groups of Myelodysplastic Syndromes). The CD34+ cells from JMML Spleens (n=5) and Bone Marrow (BM n=1) alongside healthy controls (cord blood CB n=5; BM n=2) were stained with antibodies against 15 immunophenotypic and 46 key histone markers and subsequently analyzed on a mass cytometer (Fluidigm, Helios). We found that a group of key histone markers were significantly downregulated in JMML in comparison to healthy CBs, including H4K16ac, H3K23ac, H3K56ac, H3K36me1, H3K9me1 and H3K27me2 (Fig.1). Dimensionality reduction analyses showed a global reduction in histone acetylation in all JMML spleen samples (~70%) when compared to CBs (average >95%). Our study revealed significant dysregulation of key histone markers and their respective modifiers (histone modifying enzymes) within the leukemogenic CD34+ compartment (Fig.2). Our findings demonstrated an aberrant epigenetic regulation in JMML patient samples with concurrent loss of histone acetylation and some histone methylation markers, which have been reported as hallmarks of other cancers. Further characterization of the epigenetic landscape in JMML was performed using transposase-accessible chromatin with high-throughput sequencing (bulk ATAC-seq). Notably, ATAC-seq data revealed significantly lower expression of key histone acetyltranferases (HAT), such as Kat6b and Kat8 (Fig.2) , which specifically regulate acetylation of H3K23 and H4K16 respectively. Additionally, an increased activity of histone deacetylase, HDAC9, was also observed in these samples (Fig.2). Collectively, our data suggests aberrant activities of histone modifying enzymes (HMEs) in JMML HSPCs (Fig.2), with both increased HDAC and decreased HAT activities observed in our ATAC-data. This finding is concurrent with our EpiTOF findings of global loss of histone acetylation in JMML (Fig.1A). It is known that histone modifications are easier to reverse when compared to DNA methylation, and we believe targeted inhibition of specific HDACs that we have found to be upregulated in JMML (such as HDAC9) might hold great therapeutic potential. Additionally, investigation into the impact of altered chromatin structure of JMML HSPCs on gene expression is currently ongoing using single-cell RNA-seq. In conclusion, our study identified a novel mechanism of epigenetic dysregulation in putative JMML leukemia initiating cells related to imbalanced activities of histone modifying enzymes which could potentially represent future therapeutic targets.
Bertaina: AdicetBio: Membership on an entity's Board of Directors or advisory committees; Neovii: Membership on an entity's Board of Directors or advisory committees; Cellevolve Bio: Membership on an entity's Board of Directors or advisory committees.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal